Methodology
RNA-GPT uses the pre-trained RNA-FM sequence encoder to embed RNA sequences, which are then passed through a linear projection layer. This layer learns to map the RNA embeddings to a shared representation space with natural language, enabling alignment with a backbone LLM, for which we chose Meta’s Llama-3 8B model. The training process is divided into two stages:
- Sequence and Modality Alignment: RNA and natural language representations are aligned.
- Instruction Tuning: The model is fine-tuned for task-specific QA generation.
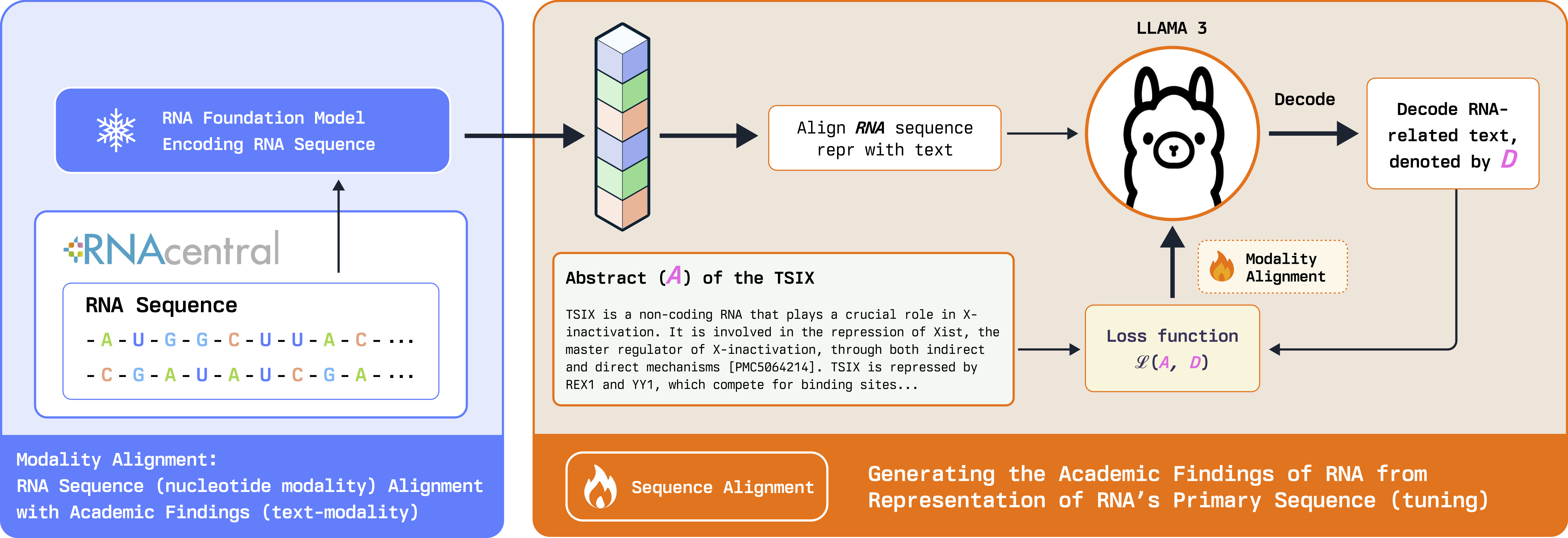
Figure 1: RNA-GPT Modality Fusion & Alignment Stage: we freeze the sequence encoder block and train the linear projection layer to learn how to align RNA sequence representations with text. In the alignment stage, the input to the training is only the projected RNA representation. No text prompts are incorporated in this stage.
Modality Alignment Stage (Stage 1): RNA sequences in the form of strings are first fed into the pre-trained sequence encoder, featuring 12 transformer layers trained with 23 million RNAs from the RNA Central database via self-supervised learning. We utilize a specialized token <RNAHere> for RNA-text modality alignment.
Instruction Tuning Stage (Stage 2): In stage 2, we instruction-tune the model using our curated RNA-QA dataset. We break down the full annotations into targeted QA samples with concise answers to specific questions as prediction targets. This allows the chat model to provide more relevant and accurate responses.
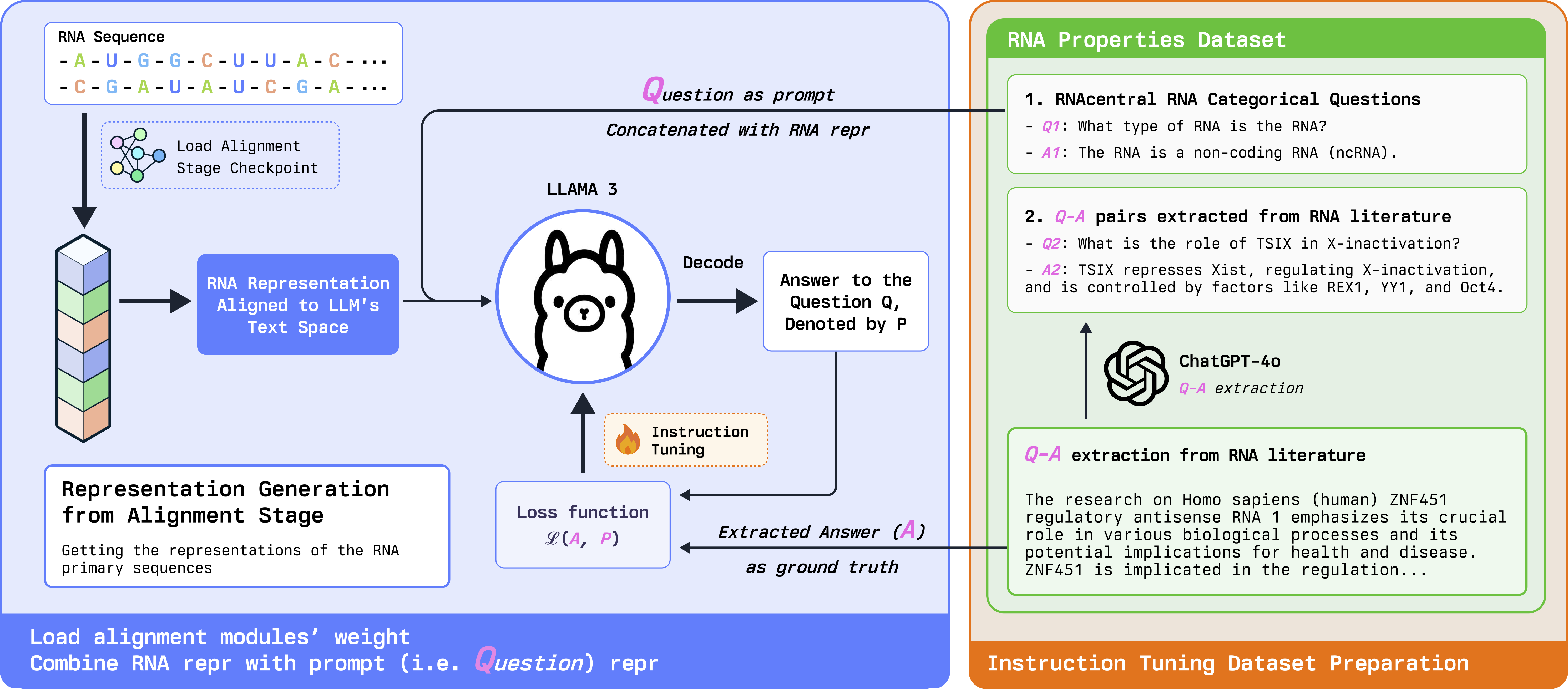
Figure 2: RNA-GPT Instruction Tuning Stage: we use the RNA representation from the alignment stage and combine it with question prompts for instruction tuning. The model generates answers that are concise and relevant to the questions.
RNA-QA Dataset
To achieve modality alignment, we constructed a large-scale dataset from the RNA Central database, comprising 407,616 RNA sequences paired with abstract descriptions.
Divide and Conquer RNA Literature Summarization: We begin by filtering RNA sequences from RNA Central, focusing on those indexed with "Lit Scan," yielding around 420,000 RNAs with associated research papers. For the remaining 407,616 RNAs, we scrape and extract abstracts from all relevant literature. We apply LDA topic modeling to group papers by topic, summarizing each group individually. This ensures each summarization focuses on a narrower, cohesive subject area, minimizing information loss.
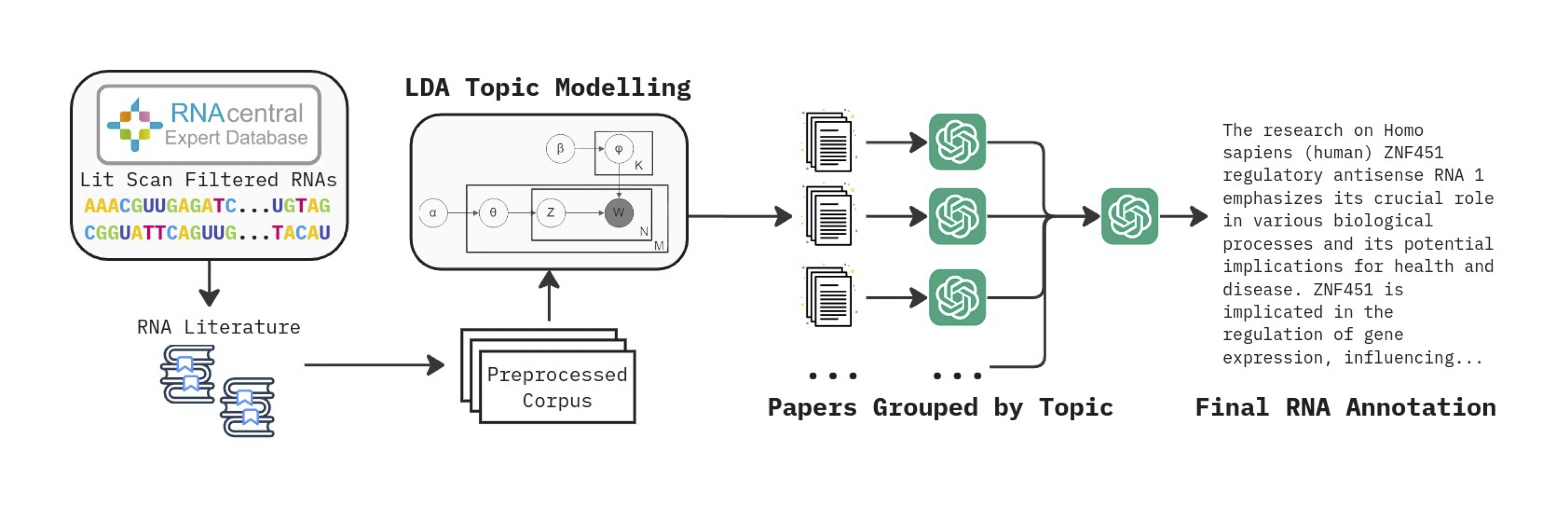
Figure 3: RNA-QA uses an automated pipeline to scrape and summarize existing RNA literature. We apply latent Dirichlet allocation (LDA) to group the vast literature on each RNA, and then we summarize each group individually using GPT-4o-mini. These summaries are then combined and refined to produce the final RNA annotation.
Data Augmentation: RNA-GPT decomposes the rich RNA annotations of RNA-QA into more specific QA-pairs for instruction tuning using GPT-4o-mini so that user instructions can be concisely answered.